Contest Problems
1.Introduction
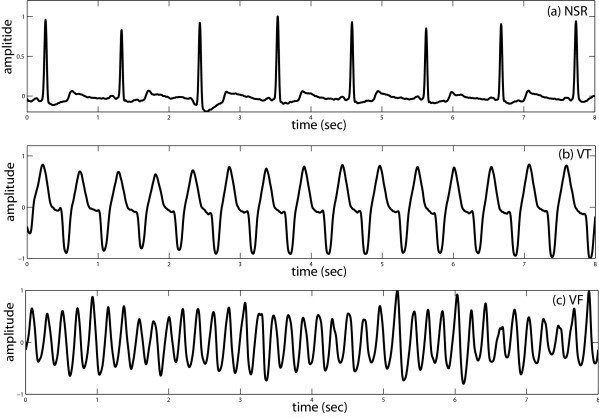
Ventricular fibrillation (VF) and ventricular tachycardia (VT) are two types of life-threatening ventricular arrhythmias (VAs),
which are the main cause of Sudden Cardiac Death (SCD). More than 60% of deaths from cardiovascular disease are from
out-of-hospital SCD [1]. It is caused by a malfunction in the heart's electrical system that can occur when the lower chambers
of the heart suddenly start beating in an uncoordinated fashion, preventing the heart from pumping blood out to the lungs and
body [1]. Unless the heart is shocked back into normal rhythm, the patient rarely survives.
People at high risk of SCD rely on the Implantable Cardioverter Defibrillator (ICD) to deliver proper and in-time defibrillation
treatment when experiencing life-threatening VAs. ICD is a small device implanted typically under the skin in the left upper
chest and is programmed to release defibrillation (i.e., electrical shock) therapy on VT and VF to restore rhythm back to
normal. The early and in-time diagnosis and treatment of life-threatening VAs can increase the chances of survival. However,
current detection methods deployed on ICDs count on a wide variety of criteria obtained from clinical trials, and there are
hundreds of programmable criteria parameters affecting the defibrillation delivery decision [2, 3].
Automatic detection with less expertise involved in ICDs can further improve the detection performances and reduce the workload from physicians in criteria design and parameters adjustment [4-6]. This year’s Challenge is asking you to build an AI/ML algorithm that can classify life-threatening VAs with only the labeled one-channel intracardiac electrograms (IEGMs) sensed by single-chamber ICDs while satisfying the requirements of in-time detection on the resource-constrained microcontroller (MCU) platform. We will test each algorithm on databases of IEGMs in terms of detection performances as well as practical performances, and the comprehensive performances will reveal the utility of the algorithm on life-threatening VAs detection in ICDs.

2.Objective
The goal of the 2022 Challenge is to discriminate life-threatening VAs (i.e., Ventricular Fibrillation and Ventricular Tachycardia) from single-lead (RVA-Bi) IEGM recordings.
We ask participants to design and implement a working, open-source AI/ML algorithm that can automatically discriminate life-threatening VAs (i.e., binary classification: VAs or non-VAs) from IEGM recordings while being able to be deployed and run on the given MCU platform. We will award prizes to the teams with top comprehensive performances in terms of detection precision, memory occupation, and inference latency.
3.Data
The data contains single-lead IEGM recordings retrieved from the lead RVA-Bi of ICD. Each recording is 5-second in length with 250 Hz sampling rate. The recordings are pre-processed by applying a band-pass filter with a frequency of a pass-band frequency of 15 Hz and a stop-band frequency of 55 Hz. Each IEGM recording has one label that describes the cardiac abnormalities (or normal sinus rhythm). Here, for the classification of VAs, it contains the labels including VT (Ventricular Tachycardia), VFb (Ventricular Fibrillation), VFt (Ventricular Flutter). For the non-VAs, the segments are labeled with the one other than VT, VFb, VFt. We have provided the lists of labels for references.
The IEGM recordings are partitioned patient-wisely in the training and testing set. 85% of the subjects’ IEGM recordings are released as training material. The rest 15% of the subjects’ IEGM recordings will be utilized to evaluate the detection performances of the submitted algorithm. The dataset for final evaluation will remain private and will not be released.
4.Data Format
All data is formatted in text. Each text file contains a 5-second IEGM recording with the sampling point on each row of 1,250 in total length. The value of each sampling point is stored as a floating-point number.
The name of each text file contains the patient ID, labels, and index separated by a dash (i.e., ”-”). For example, for the file “S001-VT-1.txt”, “S001” represents the subject number, “VT” represents the type of arrhythmias or rhythm (refer to the provided label list), “1” represents the index of the segments.
5.MCU Platform and Cube.AI
The development board required in the Challenge is NUCLEO-L432KC, which is with ARM Cortex-M4 core at 80 MHz, 258 Kbytes of flash memory, 64 Kbytes of SRAM, and embedded ST-LINK/V2-1 debugger/programmer. The development board also supports STM32 X-Cube-AI, which is an STM32Cube Expansion Package part of the STM32Cube. AI ecosystem and extending STM32CubeMX capabilities with automatic conversion of pre-trained Artificial Intelligence algorithms, including Neural Network and classical Machine Learning models, and integration of generated optimized library into the user's project. The practical performances of the AI/ML learning algorithm will be deployed and evaluated on the development board with the help of X-Cube-AI. Note that you do not have to utilize the tool X-Cube-AI to implement the project on NUCLEO-L432KC. But we will need to be able to evaluate your model performance in a straightforward way.
The board only costs around $10 and we anticipate most participating teams can easily get them online. However, if you do need support, please contact us.
Purchase Link (mouser)
Purchase Link (digikey)
6.Scoring
We will evaluate the submitted algorithm with the scoring metric that evaluates the comprehensive performances in terms of detection performances and practical performances. It is defined as follows:
For detection performances,
- score: We will compute the confusion matrix of the classification generated by the submitted algorithm on the testing dataset. The case positive is VAs. We then calculate score where with =2. This setting gives a higher weight to recall since the detection accuracy on life-threatening VAs is the most important metric in ICDs. It is expected to discriminate as many VAs recordings as possible to avoid missed detection on VAs while achieving a high detection accuracy on non-VA recordings to reduce inappropriate shock rate.
For practical performances,
- Inference latency: The average latency (in ms) of inference executed on NUCLEO-L432KC over recordings from testing dataset will be measured. The latency score will be normalized by , where and .
- Memory occupation: The memory occupation (in KiB) will be measured based on Flash occupation of NUCLEO-L432KC for the storage of the AI/ML model and the program. To be more specific, is Code+RO Data+RW Data reported by Keil when building the project. will be normalized by , where and .
The final score will be calculated as follows:
,
where the higher score is expected.
7.Example Code
We have provided an example algorithm to illustrate how to train the model with PyTorch and evaluate the comprehensive performances in terms of detection performance, flash occupation and latency.
8.References
[1] Adabag, A.S., Luepker, R.V., Roger, V.L. and Gersh, B.J., 2010. Sudden cardiac death: epidemiology and risk factors. Nature Reviews Cardiology, 7(4), pp.216-225.
[2] Zanker, N., Schuster, D., Gilkerson, J. and Stein, K., 2016. Tachycardia detection in ICDs by Boston Scientific. Herzschrittmachertherapie+ Elektrophysiologie, 27(3), pp.186-192.
[3] Madhavan, M. and Friedman, P.A., 2013. Optimal programming of implantable cardiac-defibrillators. Circulation, 128(6), pp.659-672.
[4] Jia, Z., Wang, Z., Hong, F., Ping, L., Shi, Y. and Hu, J., Learning to Learn Personalized Neural Network for Ventricular Arrhythmias Detection on Intracardiac EGMs. International Joint Conferences on Artificial Intelligence Organization (IJCAI-21), pp. 2606-2613.
[5] Acharya, U. Rajendra, et al. "Automated identification of shockable and non-shockable life-threatening ventricular arrhythmias using convolutional neural network." Future Generation Computer Systems 79 (2018): 952-959.
[6] Hannun, Awni Y., et al. "Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network." Nature medicine 25.1 (2019): 65-69.